The goal of the paper is to develop a computational microscopy technique to be able to quantitatively characterize thick biological samples. More specifically, the authors aim to obtain 3D cellular information, such as phase and absorption, without having to stain, label, or scan the sample.
Using a standard brightfield microscope, biological cells cannot be imaged, as they appear transparent when left unstained. However, the process of staining or labeling cells, which involves the injection of dyes and fluorophores, might exert ill effects on cellular functions. Therefore, a different imaging method is needed in order to reconstruct the internal 3D structure of cells.
3D phase microscopy techniques fall into two categories: interferometry-based or intensity-only. The most commonly used technique from the former category is optical diffraction tomography (ODT), in which a sample is illuminated using various angles of incidence to directly detect the phase and amplitude information of the scattered field. A reconstruction algorithm is used to retrieve the map of the 3D refractive index (RI) distribution to reveal the important optical properties of transparent samples. Nevertheless, this approach is costly due to the need for interferometry and therefore expensive and specialized equipment, which is why the second category, intensity-only phase tomography, is of more interest.
Intensity diffraction tomography (IDT), as Ling et al. describes, employs tomographic phase reconstruction from intensity-only measurements. Previous IDT methods, however, require mechanical scanning, thus increasing acquisition time and data size, result in lower spatial resolution, or necessitate an iterative reconstruction algorithm, which can be computationally intensive. For high-throughput applications, Ling et al. derive a novel technique that is able to overcome all these limitations by employing a motion-free, scan-free, and label-free IDT model that employs a linear forward reconstruction algorithm, has a slice-based framework, and utilizes angled illumination that enables the direct inversion of 3D phase and absorption from intensity-only measurements for weakly scattering samples.
The setup consists of a standard commercial microscope modified with an LED array source in which images are captured with varying illumination angles. From the 2D slices, sliced along the axial direction, the slice-wise phase and absorption transfer functions (TF) are derived at varying depths for each angle of illumination. Next, the reconstruction algorithm performs 3D synthetic aperture with slice-wise deconvolution to produce two 3D stacks, which represent the phase and absorption.
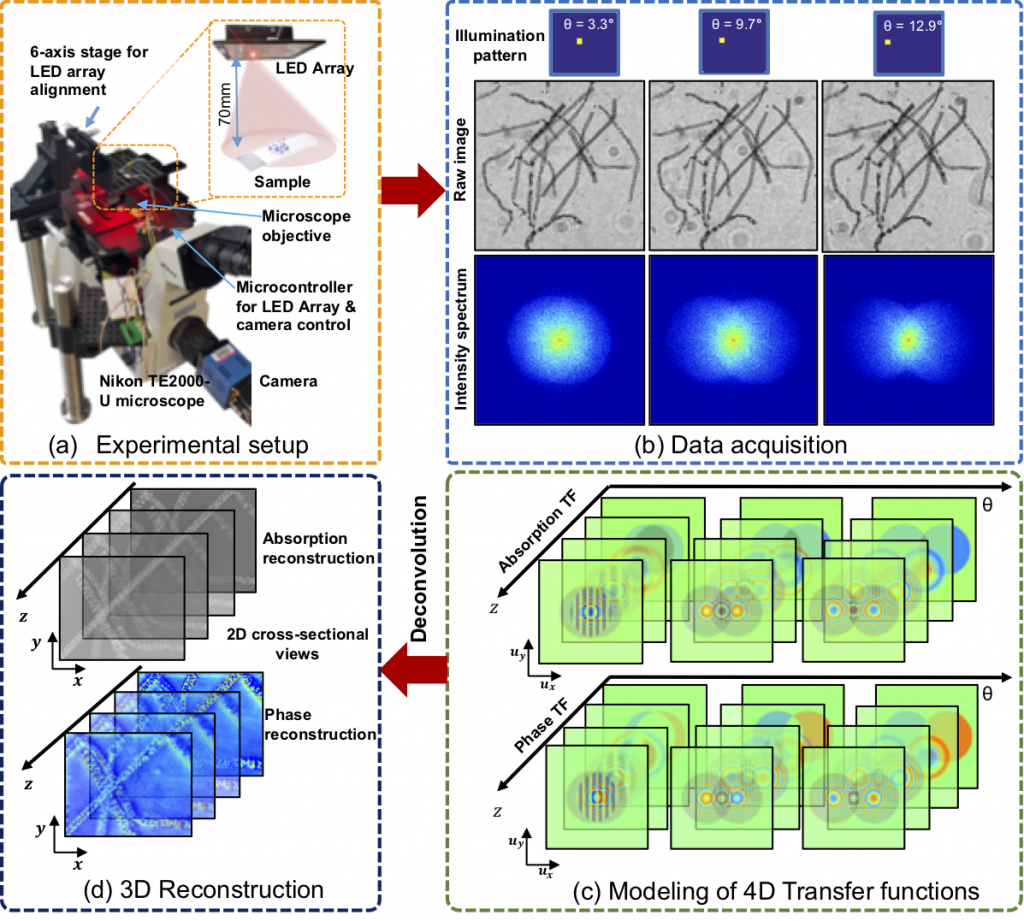
Above, a graphic from Ling et al.’s paper illustrates the experimental setup, data acquisition, 3D reconstruction, and modeling of 4D TFs.
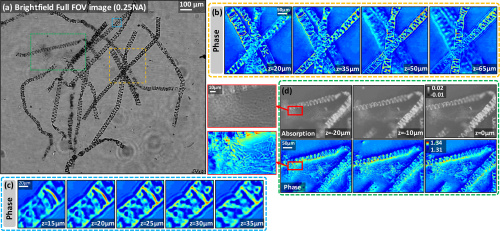
Ling et al. further validate their technique experimentally with dense biological samples including a stained spirogyra sample and unstained MCF-7 cancer cells. With the imaging of the stained 3D sample, as shown below, the authors used all of 89 brightfield images to perform the IDT reconstruction and successfully demonstrate the attainment of axial sectioning.
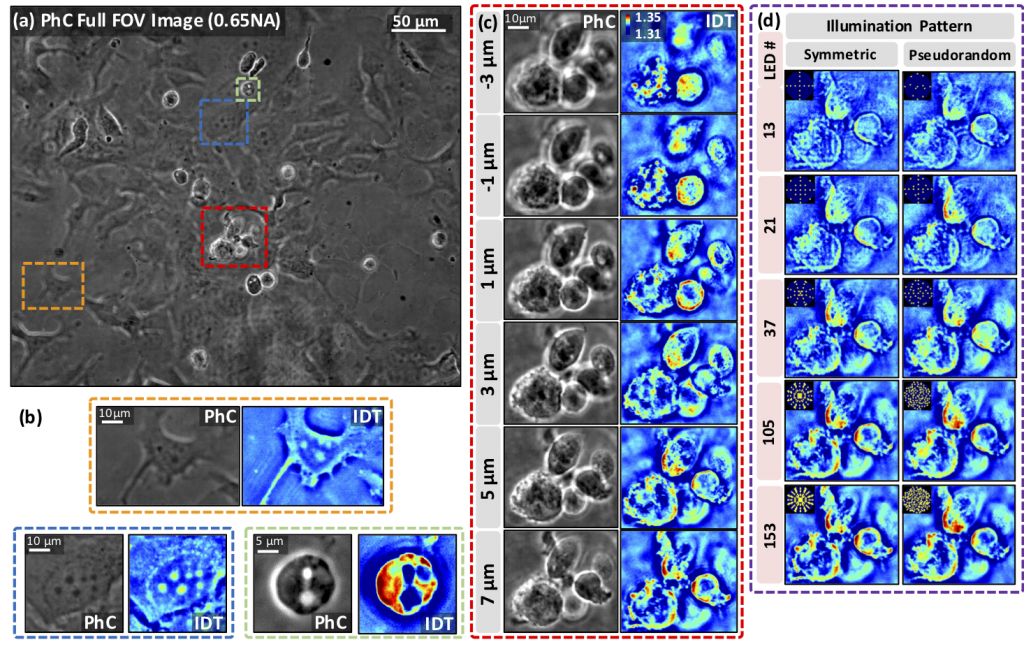
With the imaging of unstained dense cell clusters, as shown below, the authors use 153 images to reconstruct 22 phase and absorption slices and establish the flexibility and robustness of their technique in reconstructing both dense and thin samples.
Since the model and the first two experiments only account for single scattering, the authors perform a simulation and an experiment to study the effects of multiple scattering, consisting of stacked phase and absorption resolution targets, which are found to be in agreement with each other and with the theory.
In essence, this new framework is shown by the authors to allow for flexible illumination patterning, fast data acquisition, efficient memory, and low computational complexity. Moreover, this system can also be widely adopted by existing microscopy facilities given its potential for broad application with minimal hardware additions or adjustments. This is of particular interest to our lab, given the method’s capability of biological sample reconstruction and may pave the way for many more biomedical imaging applications.
References:
Ruilong Ling, Waleed Tahir, Hsing-Ying Lin, Hakho Lee, and Lei Tian, “High-throughput intensity diffraction tomography with a computational microscope,” Biomed. Opt. Express 9, 2130-2141 (2018).