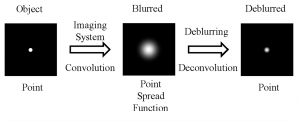
Liquid lenses are a helpful technology to overcome depth of field. Many cameras are often limited by the aperture size and resolution of the lens system, however with continuous focusability over a specified focal range, tunable liquid lenses allow for custom-tuning and optimal performance. Applications of tunable lenses include machine vision inspection systems, unmanned aerial vehicles, and measurement and dimensional rendering.
There are two main types of liquid lenses: focus-tunable lenses and variable focus liquid lenses. Focus-tunable lenses consist of a liquid optical-grade polymer that is mounted in a robust casing and is protected by two windows. These focus-tunable lenses can be either manually-focused by rotating the outer ring and manipulating the focal length, or optical power, which changes the shape from convex to concave, or they can be electrically-focused by applying a control voltage and changing the lens’ radius, which adjusts the focal length within a range of +50mm to +120mm. Variable focus liquid lenses operate via a slightly different process, called electrowetting, which is the use of electric fields to control the wetting properties by applying a voltage to increase the overall curvature, shape, and optical power of the liquid lens. Nevertheless, variable focus liquid lenses are not ideal to work with, since they are small and difficult to integrate with existing objectives., which is why I will focus on the former in this post.
Electrically-focused tunable lenses (Figure 1) provide a single-lens solution for focus and zoom objectives and eliminates the need for a multi-lens system. The lens costs between $300 and $500 and according to Edmund Optics, producer of optics and imaging technology, this device can replace the functionality of a complete lens kit in the laboratory setting. The lenses quickly adjust focus to accommodate objects located at differing working distances and thus are an ideal innovation for imaging applications that require rapid focusing, high throughput, and depth of field.
As an example, in Stay Focused! Auto Focusing Lenses for Correcting Presbyopia, Elias Wu develops active glasses for attenuating presbyopia, far-sightedness that causes the loss of the eye’s ability to focus actively on nearby objects, by employing auto-focusing lenses (ie. focus-tunable lenses) alongside a calibration-free eye-tracking method (ie. TrackingNet). The setup consists of a ski goggle-like headset with an adjustable lens holder and adjustable eye tracking mount. This device takes in data from the eye tracking cameras and runs it through TrackingNet, a deep convolutional neural network, that implements pretrained models and a multi-layer fully-connected module. This software helps to streamline the auto-focusing lens procedure.
The goal of the paper is threefold: stabilizing the frame and mount so that users could wear it securely, rendering the lens and camera within the frame adjustable, and managing the cables needed to properly operate the headset. In these regards, Wu reports that overall, the headset satisfies these three criteria, however, in regards to the software, it does not yield the best results, which may be attributed to the use of the MSE loss function, when a simple L2 loss may be better. Moreover, the ResNet-50 features convolutional layers that are optimized for classification and thus may not be suited for this type of application in which the gaze position from images of the eyes needs to be found. Most notably, Wu found that the fluid-filled tunable lenses are quite bulky and also have visible distortion around the edges of the lens (Figure 2).
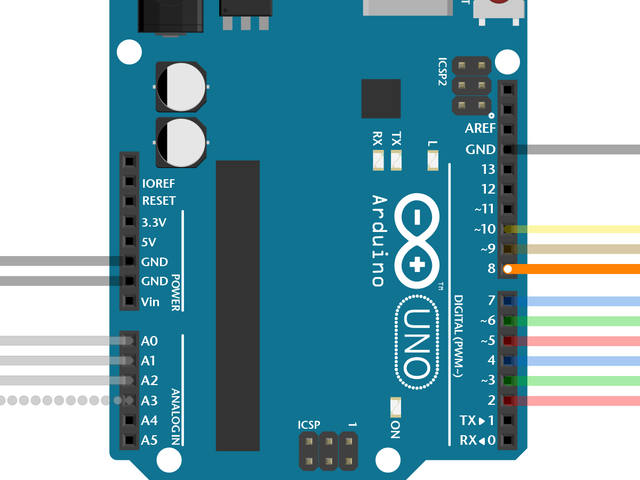
Figure 1: Focus-tunable liquid lenses (Edmund Optics)
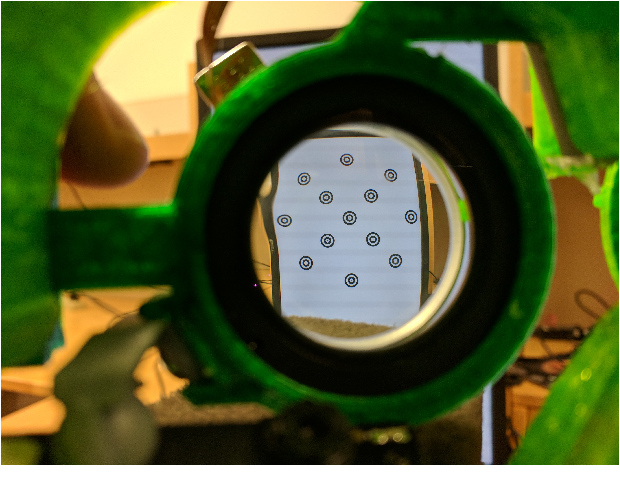
Figure 2: Distortion along edges of auto-focusing lenses (Elias Wu)