We have written a program for the dome that randomly illuminates a fixed number of LEDs at random brightness. Multiplexed illumination is useful in Fourier ptychography because it reduces data collection time without sacrificing information (X). Our code generates several patterns of LEDs and takes one photo for each pattern.
Within the program, we define the number of LEDs to be lit, the number of bright field LEDs, and the number of total LEDs. Currently, we are only illuminating bright field LEDs during imaging. We define the total number of LEDs in anticipation of future imaging that does include the dark field images.
First, we determine the number of patterns to generate by dividing the number of LEDs in use by the number to be lit in each pattern (n). We generate a list of all possible LED indices and shuffle to randomize selection. Then, we assign n indices to each pattern. We pop each index off of the shuffled list, so no index is reused. We use numpy’s randint function to generate a brightness between 0 and 255 for each index.
The inbuilt LED dome commands do not provide for individual LED brightness assignment. Instead, we must reassign the brightness of the entire array before illuminating the desired index. In order to achieve LED patterns that contain many brightnesses, we reset the dome brightness before each LED is illuminated.
We collect and save an image before wiping the current pattern to begin illuminating the next. The section of code used to save the image is not included in the interest of brevity.
for i in range(numPatterns):
for j in range(len(patterns[i])):
#Get index of current LED
light = patterns[i][j]
#Get brightness of current LED
intensity = brightness[i][j]
#Set brightness and illuminate LED
led_array.command(intensity)
led_array.command(light)
#Once the full pattern is illuminated, capture a photo
time.sleep(0.1)
mmc.snapImage()
#Clear the array in anticipation of the next pattern
led_array.command('x')
Interfacing with the dome occurs extremely quickly. It is reasonable to reset the dome brightness and individually illuminate each LED because, even using this more tedious method, the entire pattern takes only a fraction of a second to display.
The LEDs in our dome are extremely bright, so we are able to significantly reduce the camera exposure time. Previously, we used an exposure of around 2 seconds for illumination by a single LED. For five LEDs of mixed brightness in our new array , an exposure of 0.15 seconds is perfectly sufficient. We have not yet determined the ideal exposure time for an image captured with only one fully bright LED illuminated.





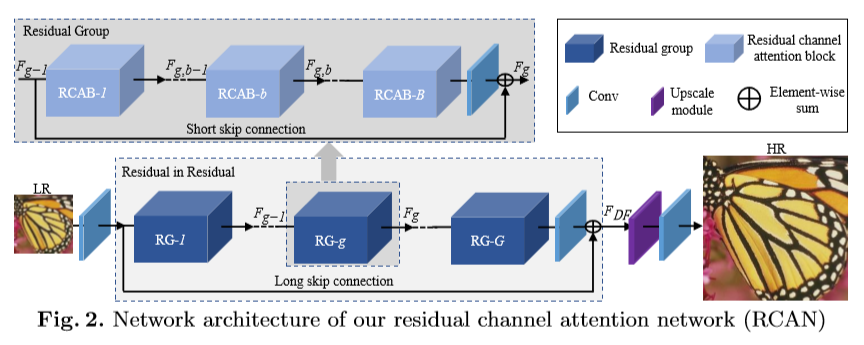
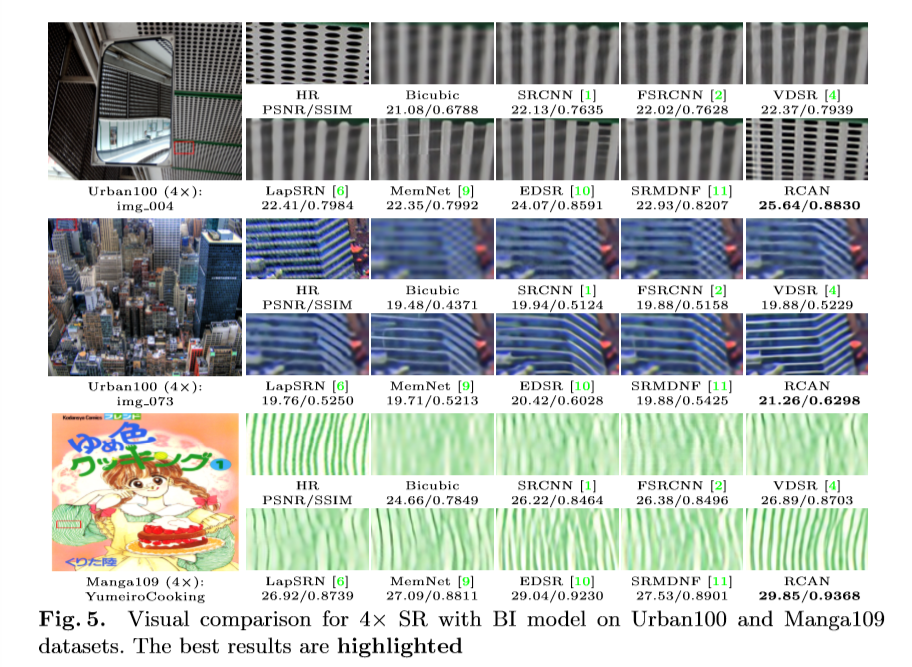


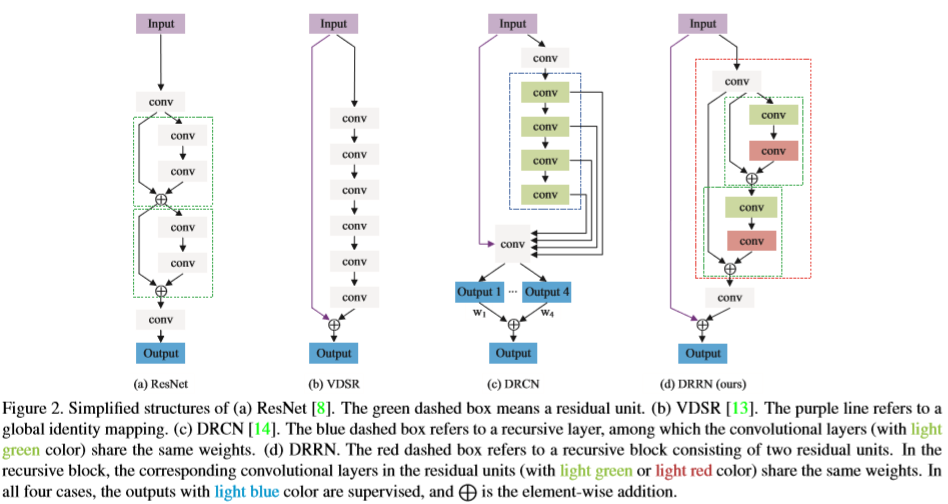
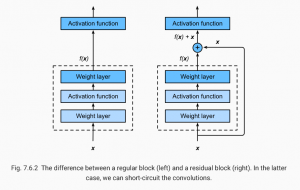 In the picture above (from the open source textbook ‘Dive into Deep Learning’), the difference between a regular (on the left) and residual block (right) is shown. The mapping output generated by the regular block, f(x), contains x more information than the residual block, and thus requires more computations. Residual blocks simplify the amount of input required and thus reduce computations.
In the picture above (from the open source textbook ‘Dive into Deep Learning’), the difference between a regular (on the left) and residual block (right) is shown. The mapping output generated by the regular block, f(x), contains x more information than the residual block, and thus requires more computations. Residual blocks simplify the amount of input required and thus reduce computations.